Research Progress
Since the outbreak of novel coronavirus 2019 (COVID-19), the number of confirmed cases has reached 1,016,000 worldwide and the global death has exceeded 53,000. This highly infectious disease is caused by the virus SARS-CoV-2. COVID-19 poses a great threat to human activities and keeps the world in suspense. A comprehensive knowledge of this novel virus will help fight against COVID-19. Several important proteins play a crucial role in the process of the virus infection. Structural information of these proteins will be helpful for the understanding of the infectious mechanism and offer a basis for the drug development. Macromolecular X-Ray Crystallographys using synchrotron radiation facility is an efficient technique to determine the 3-D structures of protein.
To meet the urgent needs of determining the protein structure, Shanghai Synchrotron Radiation Facility (SSRF) has opened a green channel to fully support the research teams carrying out studies related to COVID-19. At the end of January, SSRF promptly rebooted the accelerator that was shut down for maintenance. Three protein crystallography beamlines reopened: BL17U1, BL18U1 and BL19U1, which also played a significant role in the studies of the previous epidemic like H1N1, H7N9, MERS, Zika, and Ebola.
Since January 2020, SSRF has assisted research teams by arranging enough beam shifts (Jan. 12, Feb. 2-3, Feb. 11, Feb. 17, and Feb. 23) to carry out their studies on protein structure related to COVID-19 and selection of candidate active substances, including Zihe Rao team for the viral main protease (Mpro or also 3CLpro), Jianxun Qi and Xinquan Wang teams for the spike protein, and other teams for the drug screening.
1. On January 12, 2020, the research team led by Zihe Rao and Haitao Yang at Shanghai Tech University collected the diffraction data of COVID-19 coronavirus 3CL hydrolase (Mpro) and took the lead in determining its high-resolution crystal structure. Related link:
https://www.rcsb.org/news?year=2020&article=5e39e03fa5007a04a313edc3
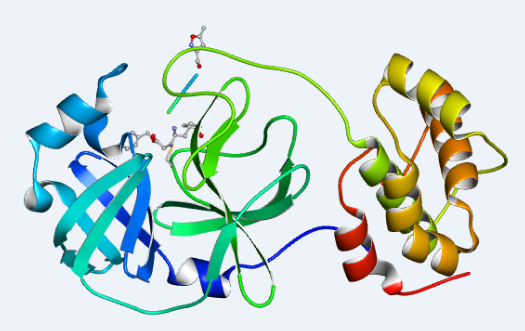
The crystal structure of COVID-19 Main protease in complex with an inhibitor N3 (PDB entry 6LU7)
2. On February 11, 2020, the research team led by Jianxun Qi at Institute of Microbiology, CAS, collected the diffraction data of COVID-19 RBD-ACE2 and shared its 2.5 ANG crystal structure to the whole community. An article titled with “structural and functional basis of SARS-CoV2 entry by using human ACE2” was published on Cell.
DOI: 10.1016/j.cell.2020.03.045
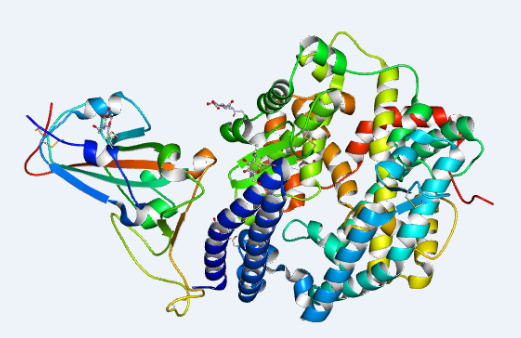
Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 (PDB entry 6LZG)
3. On February 17, 2020, the research team led by Xinquan Wang and Linqi Zhang at Tsinghua University collected the diffraction data of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, then determined and shared its crystal structure. The article was published on Natural on March 30, 2020:
https://www.nature.com/articles/s41586-020-2180-5
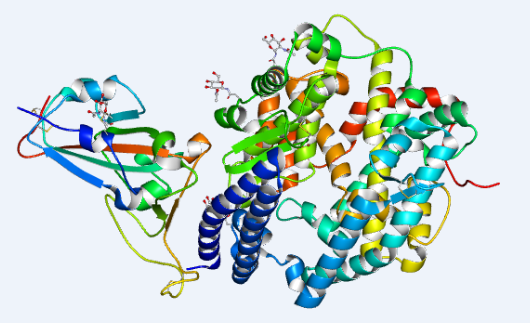
Crystal structure of COVID-19 spike receptor-binding domain bound with ACE2
(PDB entry 6M0J))
4. Moreover, beamtime was also allocated to the relevant drug screening. There are six research teams working on drug development at SSRF during this rapid access program simultaneously. Dozens of potential active substances are screened.





