Research Progress
Messenger RNA (mRNA) vaccines targeting pan-cancer therapy hold significant academic and economic value in drug research. A key challenge in mRNA design is the construction of delivery systems called lipid nanoparticles (LNPs), which serve as carriers to deliver mRNA therapies or vaccines to target cells. The preparation and screening of LNPs components involve long cycles and high costs.
Driven by this challenge, a joint research team led by Prof. LIU Lizhuang from the Shanghai Advanced Research Institute (SARI) of the Chinese Academy of Sciences proposed a deep learning model named TransLNP, which based on self-attention mechanisms that maps the three-dimensional microstructure and biochemical properties of mRNA-LNPs to enable high-precision automated screening of LNPs.
The research findings were published in Briefings in Bioinformatics.
The designed TransLNP used a cross-molecule automatic learning approach to extract knowledge from existing molecular data, enabling small-sample training for LNPs and facilitating model transfer across different molecule types.
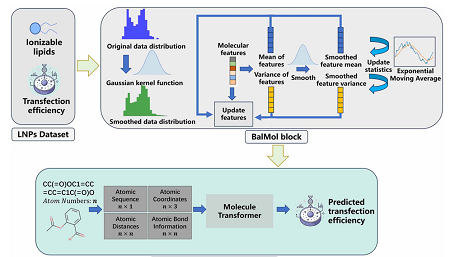
Figure 1 The overall architecture of TransLNP (Image by SARI)
To construct the mapping relationship between the 3D microstructure and biochemical properties of mRNA-LNPs, the model fully leveraged coarse-grained atomic sequence information and fine-grained atomic spatial correspondences. It extractd molecular-level features through the interaction of atomic information (atom types, coordinates, relative distance matrices, edge type matrices) based on a self-attention mechanism.
To address the imbalance caused by limited LNP data, scientists designed the BalMol (balance molecule) module. This module balanced the data by smoothing label distributions and molecular feature distributions. TransLNP achieved a mean squared error (MSE) of less than 5 for predicting LNP transfection efficiency. Compared with various mainstream graph convolutional neural networks and machine learning algorithms, TransLNP showd superior performance in terms of MSE, R2 (the larger the value, the better), and Pearson correlation coefficient, achieving top-tier metrics in the field.
This work is helpful for the rapid and accurate prediction of mRNA-LNP transfection efficiency and the prediction of new lipid nanoparticle structures, and sheds light on the application of mRNA drugs in gene therapy, vaccine development, and drug delivery.





